The Bionamic Application
The software platform for antibody discovery and development
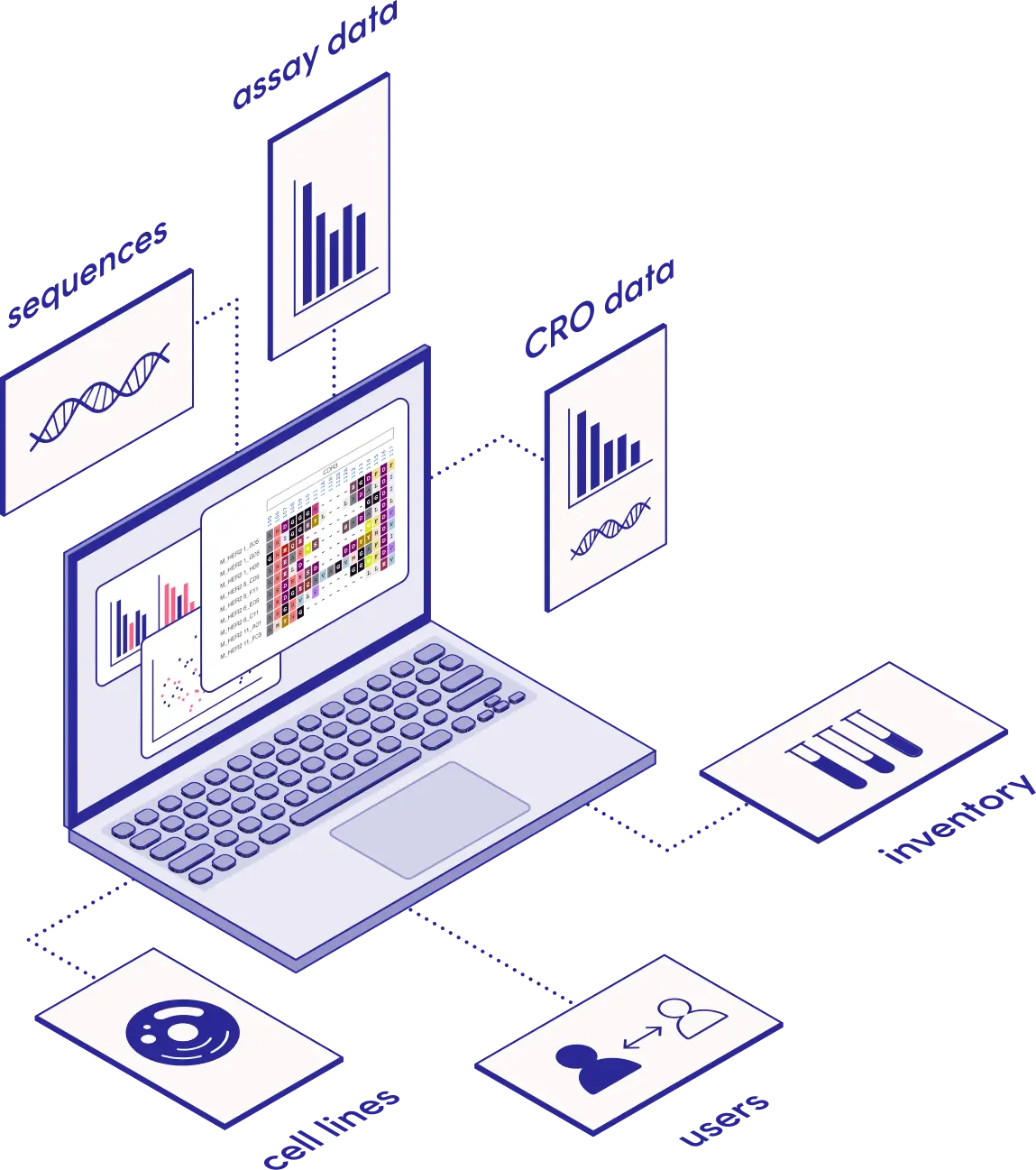
The Software Platform for Antibody Discovery and Development
- Browser based application
- Hosted or on-prem with full support
- Single source of truth for all data
- Analyze assays & annotate sequences
- Track plates, inventory and CRO data
- Powerful visualization and bioinformatics
- Automate workflows using scripts
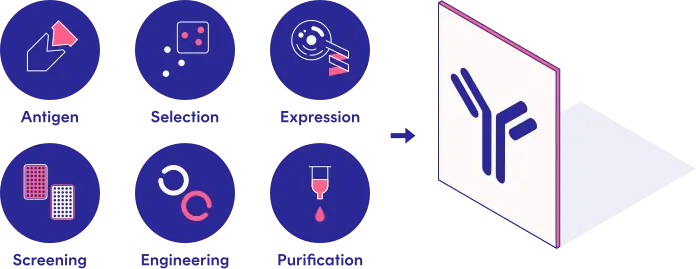
Designed for Antibody Research
- Track relations across antigens, targets and libraries
- Binder selection, screening and sequence models
- Clone new antibodies, including multispecifics and ADCs
- Find clones similar to previous hits
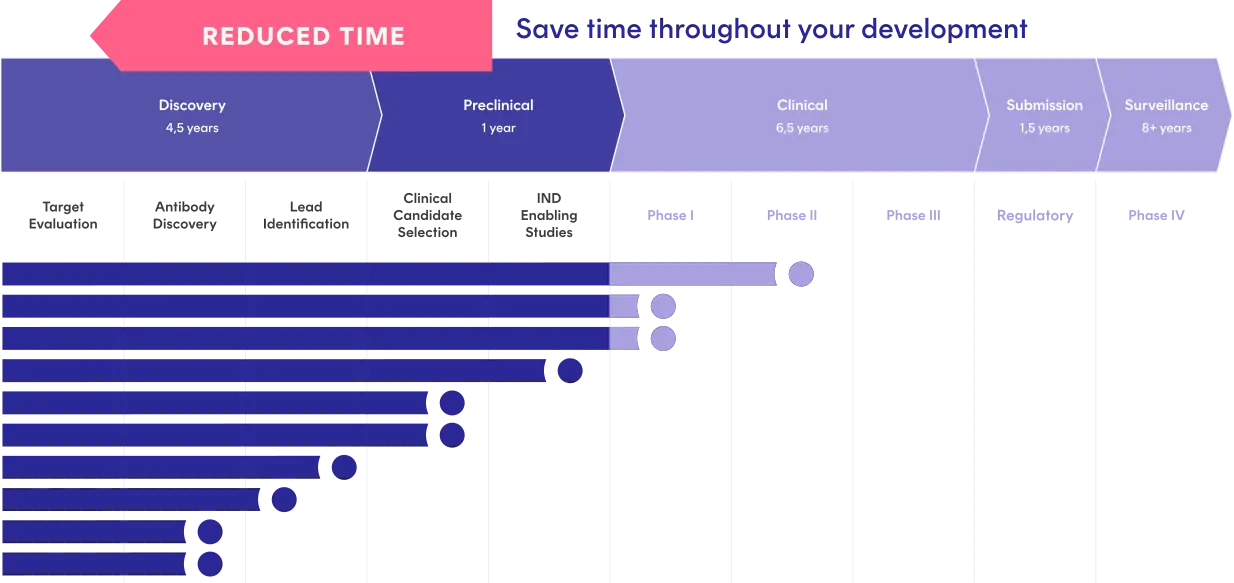
Move from Research Through Development
Within a single system, you can track all your antibody discovery and development data.
- Trace clones back to origins and pannings
- Follow constructs and formulations end-to-end
- Transition seamlessly from target to IND to CMC

Made for the Antibody Research Lab
Explore our platform and discover how Bionamic can accelerate your antibody research.